Bayesian Neural Network
We borrow this tutorial from the official Turing Docs. We will show how the explicit parameterization of Lux enables first-class composability with packages which expect flattened out parameter vectors.
Note: The tutorial in the official Turing docs is now using Lux instead of Flux.
We will use Turing.jl with Lux.jl to implement implementing a classification algorithm. Lets start by importing the relevant libraries.
# Import libraries
using Lux, Turing, CairoMakie, Random, Tracker, Functors, LinearAlgebra
# Sampling progress
Turing.setprogress!(true);Precompiling Lux...
561.0 ms ✓ ConcreteStructs
461.0 ms ✓ SIMDTypes
478.7 ms ✓ IfElse
492.4 ms ✓ Reexport
550.5 ms ✓ OpenLibm_jll
559.3 ms ✓ Future
557.5 ms ✓ CEnum
564.0 ms ✓ ManualMemory
574.1 ms ✓ ArgCheck
706.8 ms ✓ CompilerSupportLibraries_jll
714.0 ms ✓ Requires
766.1 ms ✓ Statistics
829.9 ms ✓ EnzymeCore
874.0 ms ✓ ADTypes
456.2 ms ✓ CommonWorldInvalidations
461.4 ms ✓ FastClosures
525.3 ms ✓ StaticArraysCore
622.2 ms ✓ ConstructionBase
669.9 ms ✓ JLLWrappers
580.2 ms ✓ NaNMath
782.3 ms ✓ Compat
1368.3 ms ✓ IrrationalConstants
618.4 ms ✓ Adapt
853.4 ms ✓ DocStringExtensions
904.6 ms ✓ CpuId
607.9 ms ✓ ADTypes → ADTypesEnzymeCoreExt
558.6 ms ✓ DiffResults
522.6 ms ✓ ConstructionBase → ConstructionBaseLinearAlgebraExt
530.7 ms ✓ ADTypes → ADTypesConstructionBaseExt
470.4 ms ✓ Compat → CompatLinearAlgebraExt
1148.0 ms ✓ ThreadingUtilities
504.6 ms ✓ EnzymeCore → AdaptExt
576.7 ms ✓ GPUArraysCore
1070.8 ms ✓ Static
784.9 ms ✓ Hwloc_jll
704.0 ms ✓ ArrayInterface
821.7 ms ✓ OpenSpecFun_jll
743.1 ms ✓ LogExpFunctions
2361.3 ms ✓ UnsafeAtomics
459.8 ms ✓ BitTwiddlingConvenienceFunctions
457.0 ms ✓ ArrayInterface → ArrayInterfaceGPUArraysCoreExt
735.7 ms ✓ Functors
467.1 ms ✓ ArrayInterface → ArrayInterfaceStaticArraysCoreExt
562.5 ms ✓ Atomix
1324.7 ms ✓ ChainRulesCore
2839.2 ms ✓ MacroTools
1404.3 ms ✓ CPUSummary
980.8 ms ✓ MLDataDevices
510.2 ms ✓ ADTypes → ADTypesChainRulesCoreExt
586.4 ms ✓ ArrayInterface → ArrayInterfaceChainRulesCoreExt
1915.8 ms ✓ StaticArrayInterface
717.3 ms ✓ PolyesterWeave
795.7 ms ✓ CommonSubexpressions
737.5 ms ✓ MLDataDevices → MLDataDevicesChainRulesCoreExt
2518.9 ms ✓ Hwloc
550.3 ms ✓ CloseOpenIntervals
624.4 ms ✓ LayoutPointers
1505.9 ms ✓ Optimisers
1606.9 ms ✓ LogExpFunctions → LogExpFunctionsChainRulesCoreExt
419.6 ms ✓ Optimisers → OptimisersEnzymeCoreExt
435.5 ms ✓ Optimisers → OptimisersAdaptExt
1743.1 ms ✓ Setfield
1779.8 ms ✓ DispatchDoctor
3060.7 ms ✓ SpecialFunctions
947.4 ms ✓ StrideArraysCore
443.8 ms ✓ DispatchDoctor → DispatchDoctorEnzymeCoreExt
662.8 ms ✓ DispatchDoctor → DispatchDoctorChainRulesCoreExt
746.6 ms ✓ DiffRules
747.6 ms ✓ Polyester
1240.1 ms ✓ LuxCore
456.3 ms ✓ LuxCore → LuxCoreEnzymeCoreExt
472.7 ms ✓ LuxCore → LuxCoreFunctorsExt
470.9 ms ✓ LuxCore → LuxCoreSetfieldExt
478.2 ms ✓ LuxCore → LuxCoreMLDataDevicesExt
1769.1 ms ✓ SpecialFunctions → SpecialFunctionsChainRulesCoreExt
730.0 ms ✓ LuxCore → LuxCoreChainRulesCoreExt
2832.3 ms ✓ WeightInitializers
7523.3 ms ✓ StaticArrays
1041.4 ms ✓ WeightInitializers → WeightInitializersChainRulesCoreExt
599.9 ms ✓ Adapt → AdaptStaticArraysExt
614.4 ms ✓ StaticArrays → StaticArraysStatisticsExt
618.3 ms ✓ ConstructionBase → ConstructionBaseStaticArraysExt
633.0 ms ✓ StaticArrays → StaticArraysChainRulesCoreExt
678.4 ms ✓ StaticArrayInterface → StaticArrayInterfaceStaticArraysExt
3691.3 ms ✓ ForwardDiff
843.2 ms ✓ ForwardDiff → ForwardDiffStaticArraysExt
3133.7 ms ✓ KernelAbstractions
622.4 ms ✓ KernelAbstractions → LinearAlgebraExt
691.0 ms ✓ KernelAbstractions → EnzymeExt
5040.7 ms ✓ NNlib
808.7 ms ✓ NNlib → NNlibEnzymeCoreExt
906.5 ms ✓ NNlib → NNlibForwardDiffExt
5427.9 ms ✓ LuxLib
8895.5 ms ✓ Lux
94 dependencies successfully precompiled in 33 seconds. 15 already precompiled.
Precompiling Turing...
434.2 ms ✓ IteratorInterfaceExtensions
459.0 ms ✓ NaturalSort
479.6 ms ✓ SimpleUnPack
479.1 ms ✓ UnPack
525.5 ms ✓ ScientificTypesBase
529.9 ms ✓ ExprTools
582.8 ms ✓ StatsAPI
647.1 ms ✓ ChangesOfVariables
654.9 ms ✓ PositiveFactorizations
790.2 ms ✓ AbstractFFTs
429.8 ms ✓ CommonSolve
924.7 ms ✓ Serialization
428.7 ms ✓ DataValueInterfaces
1002.1 ms ✓ FunctionWrappers
605.8 ms ✓ InverseFunctions
506.0 ms ✓ EnumX
673.7 ms ✓ SuiteSparse_jll
525.3 ms ✓ RealDot
1229.0 ms ✓ InitialValues
710.0 ms ✓ OrderedCollections
1292.7 ms ✓ Combinatorics
516.7 ms ✓ CompositionsBase
1363.5 ms ✓ FillArrays
487.8 ms ✓ PtrArrays
731.6 ms ✓ AbstractTrees
1407.7 ms ✓ OffsetArrays
507.8 ms ✓ DefineSingletons
561.9 ms ✓ IntervalSets → IntervalSetsStatisticsExt
520.6 ms ✓ InvertedIndices
660.2 ms ✓ DelimitedFiles
524.4 ms ✓ DataAPI
657.9 ms ✓ LRUCache
630.6 ms ✓ ProgressLogging
1376.4 ms ✓ RandomNumbers
522.3 ms ✓ ConstructionBase → ConstructionBaseIntervalSetsExt
527.8 ms ✓ SciMLStructures
748.2 ms ✓ LoggingExtras
866.2 ms ✓ FiniteDiff
811.9 ms ✓ Rmath_jll
1084.6 ms ✓ AxisArrays
1208.6 ms ✓ DifferentiationInterface
855.8 ms ✓ FFTW_jll
903.8 ms ✓ oneTBB_jll
1225.0 ms ✓ LogDensityProblems
954.1 ms ✓ L_BFGS_B_jll
1600.0 ms ✓ Baselet
516.6 ms ✓ TableTraits
1719.8 ms ✓ Crayons
642.3 ms ✓ StatisticalTraits
635.5 ms ✓ AbstractFFTs → AbstractFFTsChainRulesCoreExt
684.1 ms ✓ LogExpFunctions → LogExpFunctionsChangesOfVariablesExt
1441.1 ms ✓ ZygoteRules
608.8 ms ✓ RuntimeGeneratedFunctions
501.3 ms ✓ FunctionWrappersWrappers
586.8 ms ✓ InverseFunctions → InverseFunctionsDatesExt
1613.1 ms ✓ LazyArtifacts
650.1 ms ✓ LogExpFunctions → LogExpFunctionsInverseFunctionsExt
590.3 ms ✓ ChangesOfVariables → ChangesOfVariablesInverseFunctionsExt
1677.9 ms ✓ HypergeometricFunctions
632.7 ms ✓ Parameters
596.4 ms ✓ CompositionsBase → CompositionsBaseInverseFunctionsExt
650.8 ms ✓ FillArrays → FillArraysStatisticsExt
669.1 ms ✓ AliasTables
616.7 ms ✓ LeftChildRightSiblingTrees
608.2 ms ✓ OffsetArrays → OffsetArraysAdaptExt
2419.2 ms ✓ RecipesBase
702.4 ms ✓ Missings
647.8 ms ✓ LRUCache → SerializationExt
2875.9 ms ✓ StringManipulation
627.2 ms ✓ DifferentiationInterface → DifferentiationInterfaceFiniteDiffExt
922.9 ms ✓ FiniteDiff → FiniteDiffStaticArraysExt
663.6 ms ✓ DifferentiationInterface → DifferentiationInterfaceChainRulesCoreExt
1201.2 ms ✓ Libtask
1010.8 ms ✓ DifferentiationInterface → DifferentiationInterfaceStaticArraysExt
1236.9 ms ✓ Random123
822.8 ms ✓ LogDensityProblemsAD
1221.3 ms ✓ Rmath
2850.7 ms ✓ Distributed
756.0 ms ✓ LBFGSB
1220.7 ms ✓ DifferentiationInterface → DifferentiationInterfaceForwardDiffExt
2647.5 ms ✓ DataStructures
1150.3 ms ✓ Tables
730.5 ms ✓ LogDensityProblemsAD → LogDensityProblemsADADTypesExt
822.8 ms ✓ IntervalSets → IntervalSetsRecipesBaseExt
1124.1 ms ✓ MLJModelInterface
982.1 ms ✓ TerminalLoggers
1062.6 ms ✓ LogDensityProblemsAD → LogDensityProblemsADForwardDiffExt
934.6 ms ✓ SharedArrays
754.5 ms ✓ SortingAlgorithms
1180.1 ms ✓ ProgressMeter
658.6 ms ✓ LogDensityProblemsAD → LogDensityProblemsADDifferentiationInterfaceExt
1945.1 ms ✓ IntelOpenMP_jll
1094.5 ms ✓ StructArrays
1430.8 ms ✓ QuadGK
4722.6 ms ✓ Test
1534.6 ms ✓ NLSolversBase
783.9 ms ✓ ConsoleProgressMonitor
601.9 ms ✓ StructArrays → StructArraysAdaptExt
598.1 ms ✓ StructArrays → StructArraysGPUArraysCoreExt
465.4 ms ✓ InplaceOps
2596.4 ms ✓ StatsFuns
955.2 ms ✓ StructArrays → StructArraysStaticArraysExt
833.3 ms ✓ InverseFunctions → InverseFunctionsTestExt
5335.7 ms ✓ SparseArrays
1834.5 ms ✓ MKL_jll
1437.0 ms ✓ ChangesOfVariables → ChangesOfVariablesTestExt
1049.9 ms ✓ StatsFuns → StatsFunsInverseFunctionsExt
1715.9 ms ✓ SplittablesBase
4217.1 ms ✓ Accessors
929.7 ms ✓ DensityInterface
1864.9 ms ✓ AbstractFFTs → AbstractFFTsTestExt
912.1 ms ✓ ArrayInterface → ArrayInterfaceSparseArraysExt
968.9 ms ✓ WoodburyMatrices
1007.3 ms ✓ Statistics → SparseArraysExt
1030.1 ms ✓ ChainRulesCore → ChainRulesCoreSparseArraysExt
887.8 ms ✓ SuiteSparse
7623.3 ms ✓ Tracker
864.3 ms ✓ FiniteDiff → FiniteDiffSparseArraysExt
1038.1 ms ✓ FillArrays → FillArraysSparseArraysExt
659.6 ms ✓ Accessors → AccessorsStructArraysExt
1499.7 ms ✓ KernelAbstractions → SparseArraysExt
1026.2 ms ✓ DifferentiationInterface → DifferentiationInterfaceSparseArraysExt
2715.1 ms ✓ LineSearches
972.8 ms ✓ StructArrays → StructArraysSparseArraysExt
2392.5 ms ✓ StatsFuns → StatsFunsChainRulesCoreExt
916.5 ms ✓ Accessors → AccessorsTestExt
1171.7 ms ✓ Accessors → AccessorsDatesExt
1681.7 ms ✓ SparseMatrixColorings
1175.4 ms ✓ Accessors → AccessorsIntervalSetsExt
1079.2 ms ✓ BangBang
1007.6 ms ✓ Accessors → AccessorsStaticArraysExt
946.3 ms ✓ AxisAlgorithms
865.6 ms ✓ SparseInverseSubset
1213.0 ms ✓ PDMats
1571.2 ms ✓ NamedArrays
673.7 ms ✓ BangBang → BangBangChainRulesCoreExt
1970.9 ms ✓ SymbolicIndexingInterface
1007.8 ms ✓ BangBang → BangBangStaticArraysExt
675.0 ms ✓ BangBang → BangBangStructArraysExt
1631.8 ms ✓ ArrayInterface → ArrayInterfaceTrackerExt
1293.4 ms ✓ DifferentiationInterface → DifferentiationInterfaceSparseMatrixColoringsExt
2427.9 ms ✓ SciMLOperators
850.9 ms ✓ BangBang → BangBangTablesExt
1781.7 ms ✓ DifferentiationInterface → DifferentiationInterfaceTrackerExt
1680.2 ms ✓ LogDensityProblemsAD → LogDensityProblemsADTrackerExt
1213.5 ms ✓ MicroCollections
913.2 ms ✓ FillArrays → FillArraysPDMatsExt
744.3 ms ✓ SciMLOperators → SciMLOperatorsStaticArraysCoreExt
3223.9 ms ✓ StatsBase
856.5 ms ✓ SciMLOperators → SciMLOperatorsSparseArraysExt
4309.7 ms ✓ Roots
1806.9 ms ✓ Tracker → TrackerPDMatsExt
697.3 ms ✓ Roots → RootsChainRulesCoreExt
829.3 ms ✓ Roots → RootsForwardDiffExt
2755.1 ms ✓ Interpolations
2863.2 ms ✓ RecursiveArrayTools
5423.8 ms ✓ SparseConnectivityTracer
1255.9 ms ✓ RecursiveArrayTools → RecursiveArrayToolsStructArraysExt
1472.7 ms ✓ RecursiveArrayTools → RecursiveArrayToolsForwardDiffExt
1858.3 ms ✓ RecursiveArrayTools → RecursiveArrayToolsSparseArraysExt
4132.2 ms ✓ Transducers
2313.1 ms ✓ RecursiveArrayTools → RecursiveArrayToolsTrackerExt
4474.0 ms ✓ Optim
837.9 ms ✓ Transducers → TransducersAdaptExt
8790.3 ms ✓ FFTW
1979.3 ms ✓ SparseConnectivityTracer → SparseConnectivityTracerNaNMathExt
1955.1 ms ✓ SparseConnectivityTracer → SparseConnectivityTracerLogExpFunctionsExt
2281.4 ms ✓ SparseConnectivityTracer → SparseConnectivityTracerSpecialFunctionsExt
2764.2 ms ✓ SparseConnectivityTracer → SparseConnectivityTracerNNlibExt
1234.3 ms ✓ NNlib → NNlibFFTWExt
7519.5 ms ✓ ChainRules
2732.9 ms ✓ AbstractMCMC
19515.1 ms ✓ MLStyle
7093.9 ms ✓ Distributions
1085.2 ms ✓ ArrayInterface → ArrayInterfaceChainRulesExt
1452.8 ms ✓ SSMProblems
1641.9 ms ✓ AbstractPPL
1395.7 ms ✓ Distributions → DistributionsDensityInterfaceExt
1461.7 ms ✓ Distributions → DistributionsChainRulesCoreExt
1722.4 ms ✓ MCMCDiagnosticTools
1819.2 ms ✓ Distributions → DistributionsTestExt
3240.2 ms ✓ AdvancedHMC
1849.2 ms ✓ KernelDensity
1721.6 ms ✓ EllipticalSliceSampling
1951.8 ms ✓ AdvancedMH
2184.9 ms ✓ AdvancedPS
3878.0 ms ✓ Bijectors
4409.9 ms ✓ DistributionsAD
2508.3 ms ✓ AdvancedMH → AdvancedMHStructArraysExt
2607.2 ms ✓ AdvancedMH → AdvancedMHForwardDiffExt
21099.9 ms ✓ PrettyTables
2542.2 ms ✓ AdvancedPS → AdvancedPSLibtaskExt
1687.4 ms ✓ Bijectors → BijectorsForwardDiffExt
1663.6 ms ✓ DistributionsAD → DistributionsADForwardDiffExt
1714.0 ms ✓ Bijectors → BijectorsDistributionsADExt
2992.3 ms ✓ Bijectors → BijectorsTrackerExt
3020.0 ms ✓ DistributionsAD → DistributionsADTrackerExt
9427.8 ms ✓ Expronicon
3077.0 ms ✓ MCMCChains
2086.5 ms ✓ AdvancedVI
2263.5 ms ✓ AdvancedMH → AdvancedMHMCMCChainsExt
2266.1 ms ✓ AdvancedHMC → AdvancedHMCMCMCChainsExt
8901.0 ms ✓ DynamicPPL
1949.9 ms ✓ DynamicPPL → DynamicPPLForwardDiffExt
2025.9 ms ✓ DynamicPPL → DynamicPPLChainRulesCoreExt
2511.5 ms ✓ DynamicPPL → DynamicPPLMCMCChainsExt
2779.5 ms ✓ DynamicPPL → DynamicPPLZygoteRulesExt
11412.3 ms ✓ SciMLBase
1086.0 ms ✓ SciMLBase → SciMLBaseChainRulesCoreExt
2211.7 ms ✓ OptimizationBase
344.5 ms ✓ OptimizationBase → OptimizationFiniteDiffExt
693.3 ms ✓ OptimizationBase → OptimizationForwardDiffExt
2146.0 ms ✓ Optimization
12458.0 ms ✓ OptimizationOptimJL
5061.6 ms ✓ Turing
3908.6 ms ✓ Turing → TuringOptimExt
216 dependencies successfully precompiled in 67 seconds. 89 already precompiled.
Precompiling MLDataDevicesSparseArraysExt...
640.6 ms ✓ MLDataDevices → MLDataDevicesSparseArraysExt
1 dependency successfully precompiled in 1 seconds. 17 already precompiled.
Precompiling MLDataDevicesFillArraysExt...
413.4 ms ✓ MLDataDevices → MLDataDevicesFillArraysExt
1 dependency successfully precompiled in 0 seconds. 15 already precompiled.
Precompiling MLDataDevicesChainRulesExt...
794.9 ms ✓ MLDataDevices → MLDataDevicesChainRulesExt
1 dependency successfully precompiled in 1 seconds. 40 already precompiled.
Precompiling BijectorsEnzymeCoreExt...
1292.9 ms ✓ Bijectors → BijectorsEnzymeCoreExt
1 dependency successfully precompiled in 1 seconds. 79 already precompiled.
Precompiling HwlocTrees...
492.7 ms ✓ Hwloc → HwlocTrees
1 dependency successfully precompiled in 1 seconds. 10 already precompiled.
Precompiling StaticArrayInterfaceOffsetArraysExt...
430.7 ms ✓ StaticArrayInterface → StaticArrayInterfaceOffsetArraysExt
1 dependency successfully precompiled in 1 seconds. 18 already precompiled.
Precompiling MLDataDevicesTrackerExt...
1167.1 ms ✓ MLDataDevices → MLDataDevicesTrackerExt
1 dependency successfully precompiled in 1 seconds. 59 already precompiled.
Precompiling LuxLibTrackerExt...
1072.2 ms ✓ LuxCore → LuxCoreArrayInterfaceTrackerExt
3248.4 ms ✓ LuxLib → LuxLibTrackerExt
2 dependencies successfully precompiled in 4 seconds. 100 already precompiled.
Precompiling LuxTrackerExt...
2018.0 ms ✓ Lux → LuxTrackerExt
1 dependency successfully precompiled in 2 seconds. 114 already precompiled.
Precompiling DynamicPPLEnzymeCoreExt...
1755.2 ms ✓ DynamicPPL → DynamicPPLEnzymeCoreExt
1 dependency successfully precompiled in 2 seconds. 128 already precompiled.
Precompiling MLDataDevicesRecursiveArrayToolsExt...
596.0 ms ✓ MLDataDevices → MLDataDevicesRecursiveArrayToolsExt
1 dependency successfully precompiled in 1 seconds. 48 already precompiled.
Precompiling OptimizationMLDataDevicesExt...
1387.6 ms ✓ OptimizationBase → OptimizationMLDataDevicesExt
1 dependency successfully precompiled in 2 seconds. 97 already precompiled.
Precompiling CairoMakie...
621.0 ms ✓ PaddedViews
574.3 ms ✓ Scratch
567.7 ms ✓ SignedDistanceFields
596.0 ms ✓ StackViews
593.2 ms ✓ Showoff
833.0 ms ✓ LLVMOpenMP_jll
854.6 ms ✓ Graphite2_jll
851.6 ms ✓ Xorg_libXau_jll
875.7 ms ✓ Libmount_jll
876.0 ms ✓ Bzip2_jll
901.0 ms ✓ OpenSSL_jll
807.1 ms ✓ libfdk_aac_jll
806.4 ms ✓ Imath_jll
833.6 ms ✓ libpng_jll
820.6 ms ✓ Giflib_jll
1445.5 ms ✓ GeoInterface
873.1 ms ✓ LAME_jll
1547.2 ms ✓ SimpleTraits
846.7 ms ✓ LERC_jll
828.2 ms ✓ EarCut_jll
846.4 ms ✓ CRlibm_jll
834.2 ms ✓ Ogg_jll
917.6 ms ✓ XZ_jll
962.9 ms ✓ JpegTurbo_jll
812.4 ms ✓ Xorg_libXdmcp_jll
840.9 ms ✓ x265_jll
868.7 ms ✓ x264_jll
883.0 ms ✓ libaom_jll
858.3 ms ✓ Zstd_jll
2325.3 ms ✓ UnicodeFun
867.0 ms ✓ Expat_jll
737.9 ms ✓ Xorg_xtrans_jll
824.3 ms ✓ LZO_jll
838.4 ms ✓ Opus_jll
878.3 ms ✓ Libiconv_jll
851.9 ms ✓ Libffi_jll
832.3 ms ✓ Libgpg_error_jll
848.4 ms ✓ isoband_jll
852.5 ms ✓ Xorg_libpthread_stubs_jll
707.8 ms ✓ RelocatableFolders
975.3 ms ✓ FriBidi_jll
940.1 ms ✓ Libuuid_jll
843.8 ms ✓ MosaicViews
3334.9 ms ✓ FixedPointNumbers
959.2 ms ✓ FreeType2_jll
1044.9 ms ✓ Pixman_jll
1048.5 ms ✓ OpenEXR_jll
1520.9 ms ✓ FilePathsBase
610.1 ms ✓ Isoband
925.6 ms ✓ libsixel_jll
982.4 ms ✓ libvorbis_jll
547.0 ms ✓ Ratios → RatiosFixedPointNumbersExt
850.8 ms ✓ Libgcrypt_jll
928.5 ms ✓ Libtiff_jll
933.7 ms ✓ XML2_jll
709.8 ms ✓ FilePathsBase → FilePathsBaseMmapExt
866.7 ms ✓ Fontconfig_jll
1392.3 ms ✓ FreeType
913.2 ms ✓ XSLT_jll
933.0 ms ✓ Gettext_jll
1120.8 ms ✓ FilePaths
1454.4 ms ✓ FilePathsBase → FilePathsBaseTestExt
3335.7 ms ✓ IntervalArithmetic
1010.7 ms ✓ Glib_jll
1589.0 ms ✓ Xorg_libxcb_jll
696.3 ms ✓ IntervalArithmetic → IntervalArithmeticIntervalSetsExt
3342.0 ms ✓ ColorTypes
4551.7 ms ✓ PkgVersion
4885.5 ms ✓ FileIO
783.5 ms ✓ Xorg_libX11_jll
871.8 ms ✓ Xorg_libXrender_jll
873.1 ms ✓ Xorg_libXext_jll
1796.6 ms ✓ QOI
2239.2 ms ✓ ColorVectorSpace
6535.0 ms ✓ GeometryBasics
1040.5 ms ✓ Cairo_jll
1063.5 ms ✓ Libglvnd_jll
768.4 ms ✓ ColorVectorSpace → SpecialFunctionsExt
1099.2 ms ✓ HarfBuzz_jll
1189.5 ms ✓ libwebp_jll
1558.4 ms ✓ Packing
4870.9 ms ✓ ExactPredicates
1888.2 ms ✓ ShaderAbstractions
9368.8 ms ✓ SIMD
1244.4 ms ✓ libass_jll
1270.0 ms ✓ Pango_jll
4931.8 ms ✓ Colors
785.7 ms ✓ Graphics
806.3 ms ✓ Animations
1272.9 ms ✓ FFMPEG_jll
1504.9 ms ✓ ColorBrewer
2031.0 ms ✓ OpenEXR
5004.2 ms ✓ MakieCore
1824.0 ms ✓ Cairo
2664.3 ms ✓ FreeTypeAbstraction
4299.3 ms ✓ ColorSchemes
7270.9 ms ✓ GridLayoutBase
6626.5 ms ✓ DelaunayTriangulation
20938.4 ms ✓ Unitful
694.2 ms ✓ Unitful → InverseFunctionsUnitfulExt
718.9 ms ✓ Unitful → ConstructionBaseUnitfulExt
1628.2 ms ✓ Interpolations → InterpolationsUnitfulExt
11097.7 ms ✓ Automa
9704.3 ms ✓ PlotUtils
17027.6 ms ✓ ImageCore
2052.5 ms ✓ ImageBase
2582.3 ms ✓ WebP
3516.3 ms ✓ PNGFiles
3574.9 ms ✓ JpegTurbo
2139.1 ms ✓ ImageAxes
4605.6 ms ✓ Sixel
1247.1 ms ✓ ImageMetadata
12609.4 ms ✓ MathTeXEngine
2100.2 ms ✓ Netpbm
49881.9 ms ✓ TiffImages
1172.0 ms ✓ ImageIO
112998.7 ms ✓ Makie
75073.1 ms ✓ CairoMakie
118 dependencies successfully precompiled in 251 seconds. 151 already precompiled.
Precompiling SparseMatrixColoringsColorsExt...
856.4 ms ✓ SparseMatrixColorings → SparseMatrixColoringsColorsExt
1 dependency successfully precompiled in 1 seconds. 29 already precompiled.
Precompiling AccessorsUnitfulExt...
597.7 ms ✓ Accessors → AccessorsUnitfulExt
1 dependency successfully precompiled in 1 seconds. 17 already precompiled.
Precompiling IntervalArithmeticForwardDiffExt...
475.2 ms ✓ IntervalArithmetic → IntervalArithmeticDiffRulesExt
660.6 ms ✓ IntervalArithmetic → IntervalArithmeticForwardDiffExt
2 dependencies successfully precompiled in 1 seconds. 42 already precompiled.
Precompiling IntervalArithmeticRecipesBaseExt...
781.8 ms ✓ IntervalArithmetic → IntervalArithmeticRecipesBaseExt
1 dependency successfully precompiled in 1 seconds. 31 already precompiled.
Precompiling SciMLBaseMakieExt...
9300.0 ms ✓ SciMLBase → SciMLBaseMakieExt
1 dependency successfully precompiled in 10 seconds. 303 already precompiled.
[ Info: [Turing]: progress logging is enabled globally
[ Info: [AdvancedVI]: global PROGRESS is set as trueGenerating data
Our goal here is to use a Bayesian neural network to classify points in an artificial dataset. The code below generates data points arranged in a box-like pattern and displays a graph of the dataset we'll be working with.
# Number of points to generate
N = 80
M = round(Int, N / 4)
rng = Random.default_rng()
Random.seed!(rng, 1234)
# Generate artificial data
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
xt1s = Array([[x1s[i] + 0.5f0; x2s[i] + 0.5f0] for i in 1:M])
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
append!(xt1s, Array([[x1s[i] - 5.0f0; x2s[i] - 5.0f0] for i in 1:M]))
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
xt0s = Array([[x1s[i] + 0.5f0; x2s[i] - 5.0f0] for i in 1:M])
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
append!(xt0s, Array([[x1s[i] - 5.0f0; x2s[i] + 0.5f0] for i in 1:M]))
# Store all the data for later
xs = [xt1s; xt0s]
ts = [ones(2 * M); zeros(2 * M)]
# Plot data points
function plot_data()
x1 = first.(xt1s)
y1 = last.(xt1s)
x2 = first.(xt0s)
y2 = last.(xt0s)
fig = Figure()
ax = CairoMakie.Axis(fig[1, 1]; xlabel="x", ylabel="y")
scatter!(ax, x1, y1; markersize=16, color=:red, strokecolor=:black, strokewidth=2)
scatter!(ax, x2, y2; markersize=16, color=:blue, strokecolor=:black, strokewidth=2)
return fig
end
plot_data()Building the Neural Network
The next step is to define a feedforward neural network where we express our parameters as distributions, and not single points as with traditional neural networks. For this we will use Dense to define liner layers and compose them via Chain, both are neural network primitives from Lux. The network nn we will create will have two hidden layers with tanh activations and one output layer with sigmoid activation, as shown below.
The nn is an instance that acts as a function and can take data, parameters and current state as inputs and output predictions. We will define distributions on the neural network parameters.
# Construct a neural network using Lux
nn = Chain(Dense(2 => 3, tanh), Dense(3 => 2, tanh), Dense(2 => 1, sigmoid))
# Initialize the model weights and state
ps, st = Lux.setup(rng, nn)
Lux.parameterlength(nn) # number of parameters in NN20The probabilistic model specification below creates a parameters variable, which has IID normal variables. The parameters represents all parameters of our neural net (weights and biases).
# Create a regularization term and a Gaussian prior variance term.
alpha = 0.09
sig = sqrt(1.0 / alpha)3.3333333333333335Construct named tuple from a sampled parameter vector. We could also use ComponentArrays here and simply broadcast to avoid doing this. But let's do it this way to avoid dependencies.
function vector_to_parameters(ps_new::AbstractVector, ps::NamedTuple)
@assert length(ps_new) == Lux.parameterlength(ps)
i = 1
function get_ps(x)
z = reshape(view(ps_new, i:(i + length(x) - 1)), size(x))
i += length(x)
return z
end
return fmap(get_ps, ps)
endvector_to_parameters (generic function with 1 method)To interface with external libraries it is often desirable to use the StatefulLuxLayer to automatically handle the neural network states.
const model = StatefulLuxLayer{true}(nn, nothing, st)
# Specify the probabilistic model.
@model function bayes_nn(xs, ts)
# Sample the parameters
nparameters = Lux.parameterlength(nn)
parameters ~ MvNormal(zeros(nparameters), Diagonal(abs2.(sig .* ones(nparameters))))
# Forward NN to make predictions
preds = Lux.apply(model, xs, vector_to_parameters(parameters, ps))
# Observe each prediction.
for i in eachindex(ts)
ts[i] ~ Bernoulli(preds[i])
end
endbayes_nn (generic function with 2 methods)Inference can now be performed by calling sample. We use the HMC sampler here.
# Perform inference.
N = 5000
ch = sample(bayes_nn(reduce(hcat, xs), ts), HMC(0.05, 4; adtype=AutoTracker()), N)Chains MCMC chain (5000×30×1 Array{Float64, 3}):
Iterations = 1:1:5000
Number of chains = 1
Samples per chain = 5000
Wall duration = 24.4 seconds
Compute duration = 24.4 seconds
parameters = parameters[1], parameters[2], parameters[3], parameters[4], parameters[5], parameters[6], parameters[7], parameters[8], parameters[9], parameters[10], parameters[11], parameters[12], parameters[13], parameters[14], parameters[15], parameters[16], parameters[17], parameters[18], parameters[19], parameters[20]
internals = lp, n_steps, is_accept, acceptance_rate, log_density, hamiltonian_energy, hamiltonian_energy_error, numerical_error, step_size, nom_step_size
Summary Statistics
parameters mean std mcse ess_bulk ess_tail rhat ess_per_sec
Symbol Float64 Float64 Float64 Float64 Float64 Float64 Float64
parameters[1] 5.8536 2.5579 0.6449 16.6210 21.2567 1.2169 0.6811
parameters[2] 0.1106 0.3642 0.0467 76.1493 35.8893 1.0235 3.1205
parameters[3] 4.1685 2.2970 0.6025 15.9725 62.4537 1.0480 0.6545
parameters[4] 1.0580 1.9179 0.4441 22.3066 51.3818 1.0513 0.9141
parameters[5] 4.7925 2.0622 0.5484 15.4001 28.2539 1.1175 0.6311
parameters[6] 0.7155 1.3734 0.2603 28.7492 59.2257 1.0269 1.1781
parameters[7] 0.4981 2.7530 0.7495 14.5593 22.0260 1.2506 0.5966
parameters[8] 0.4568 1.1324 0.2031 31.9424 38.7102 1.0447 1.3090
parameters[9] -1.0215 2.6186 0.7268 14.2896 22.8493 1.2278 0.5856
parameters[10] 2.1324 1.6319 0.4231 15.0454 43.2111 1.3708 0.6165
parameters[11] -2.0262 1.8130 0.4727 15.0003 23.5212 1.2630 0.6147
parameters[12] -4.5525 1.9168 0.4399 18.6812 29.9668 1.0581 0.7655
parameters[13] 3.7207 1.3736 0.2889 22.9673 55.7445 1.0128 0.9412
parameters[14] 2.5799 1.7626 0.4405 17.7089 38.8364 1.1358 0.7257
parameters[15] -1.3181 1.9554 0.5213 14.6312 22.0160 1.1793 0.5996
parameters[16] -2.9322 1.2308 0.2334 28.3970 130.8667 1.0216 1.1637
parameters[17] -2.4957 2.7976 0.7745 16.2068 20.1562 1.0692 0.6641
parameters[18] -5.0880 1.1401 0.1828 39.8971 52.4786 1.1085 1.6349
parameters[19] -4.7674 2.0627 0.5354 21.4562 18.3886 1.0764 0.8792
parameters[20] -4.7466 1.2214 0.2043 38.5170 32.7162 1.0004 1.5784
Quantiles
parameters 2.5% 25.0% 50.0% 75.0% 97.5%
Symbol Float64 Float64 Float64 Float64 Float64
parameters[1] 0.9164 4.2536 5.9940 7.2512 12.0283
parameters[2] -0.5080 -0.1044 0.0855 0.2984 1.0043
parameters[3] 0.3276 2.1438 4.2390 6.1737 7.8532
parameters[4] -1.4579 -0.1269 0.4550 1.6893 5.8331
parameters[5] 1.4611 3.3711 4.4965 5.6720 9.3282
parameters[6] -1.2114 -0.1218 0.4172 1.2724 4.1938
parameters[7] -6.0297 -0.5712 0.5929 2.1686 5.8786
parameters[8] -1.8791 -0.2492 0.4862 1.1814 2.9032
parameters[9] -6.7656 -2.6609 -0.4230 0.9269 2.8021
parameters[10] -1.2108 1.0782 2.0899 3.3048 5.0428
parameters[11] -6.1454 -3.0731 -2.0592 -1.0526 1.8166
parameters[12] -8.8873 -5.8079 -4.2395 -3.2409 -1.2353
parameters[13] 1.2909 2.6693 3.7502 4.6268 6.7316
parameters[14] -0.2741 1.2807 2.2801 3.5679 6.4876
parameters[15] -4.7115 -2.6584 -1.4956 -0.2644 3.3498
parameters[16] -5.4427 -3.7860 -2.8946 -1.9382 -0.8417
parameters[17] -6.4221 -4.0549 -2.9178 -1.7934 5.5835
parameters[18] -7.5413 -5.8069 -5.0388 -4.3025 -3.0121
parameters[19] -7.2611 -5.9449 -5.2768 -4.3663 2.1958
parameters[20] -7.0130 -5.5204 -4.8727 -3.9813 -1.9280Now we extract the parameter samples from the sampled chain as θ (this is of size 5000 x 20 where 5000 is the number of iterations and 20 is the number of parameters). We'll use these primarily to determine how good our model's classifier is.
# Extract all weight and bias parameters.
θ = MCMCChains.group(ch, :parameters).value;Prediction Visualization
# A helper to run the nn through data `x` using parameters `θ`
nn_forward(x, θ) = model(x, vector_to_parameters(θ, ps))
# Plot the data we have.
fig = plot_data()
# Find the index that provided the highest log posterior in the chain.
_, i = findmax(ch[:lp])
# Extract the max row value from i.
i = i.I[1]
# Plot the posterior distribution with a contour plot
x1_range = collect(range(-6; stop=6, length=25))
x2_range = collect(range(-6; stop=6, length=25))
Z = [nn_forward([x1, x2], θ[i, :])[1] for x1 in x1_range, x2 in x2_range]
contour!(x1_range, x2_range, Z; linewidth=3, colormap=:seaborn_bright)
figThe contour plot above shows that the MAP method is not too bad at classifying our data. Now we can visualize our predictions.
The nn_predict function takes the average predicted value from a network parameterized by weights drawn from the MCMC chain.
# Return the average predicted value across multiple weights.
nn_predict(x, θ, num) = mean([first(nn_forward(x, view(θ, i, :))) for i in 1:10:num])nn_predict (generic function with 1 method)Next, we use the nn_predict function to predict the value at a sample of points where the x1 and x2 coordinates range between -6 and 6. As we can see below, we still have a satisfactory fit to our data, and more importantly, we can also see where the neural network is uncertain about its predictions much easier–-those regions between cluster boundaries.
Plot the average prediction.
fig = plot_data()
n_end = 1500
x1_range = collect(range(-6; stop=6, length=25))
x2_range = collect(range(-6; stop=6, length=25))
Z = [nn_predict([x1, x2], θ, n_end)[1] for x1 in x1_range, x2 in x2_range]
contour!(x1_range, x2_range, Z; linewidth=3, colormap=:seaborn_bright)
figSuppose we are interested in how the predictive power of our Bayesian neural network evolved between samples. In that case, the following graph displays an animation of the contour plot generated from the network weights in samples 1 to 5,000.
fig = plot_data()
Z = [first(nn_forward([x1, x2], θ[1, :])) for x1 in x1_range, x2 in x2_range]
c = contour!(x1_range, x2_range, Z; linewidth=3, colormap=:seaborn_bright)
record(fig, "results.gif", 1:250:size(θ, 1)) do i
fig.current_axis[].title = "Iteration: $i"
Z = [first(nn_forward([x1, x2], θ[i, :])) for x1 in x1_range, x2 in x2_range]
c[3] = Z
return fig
end"results.gif"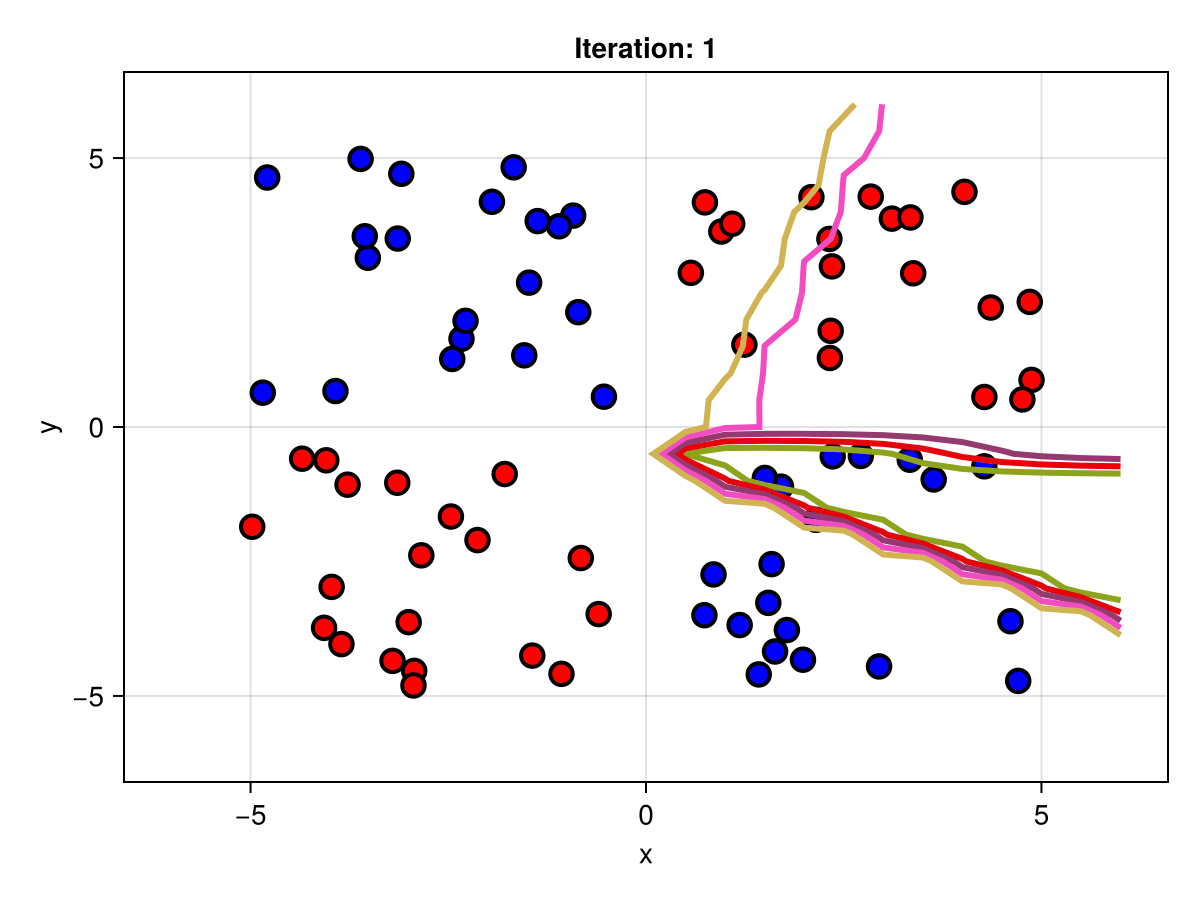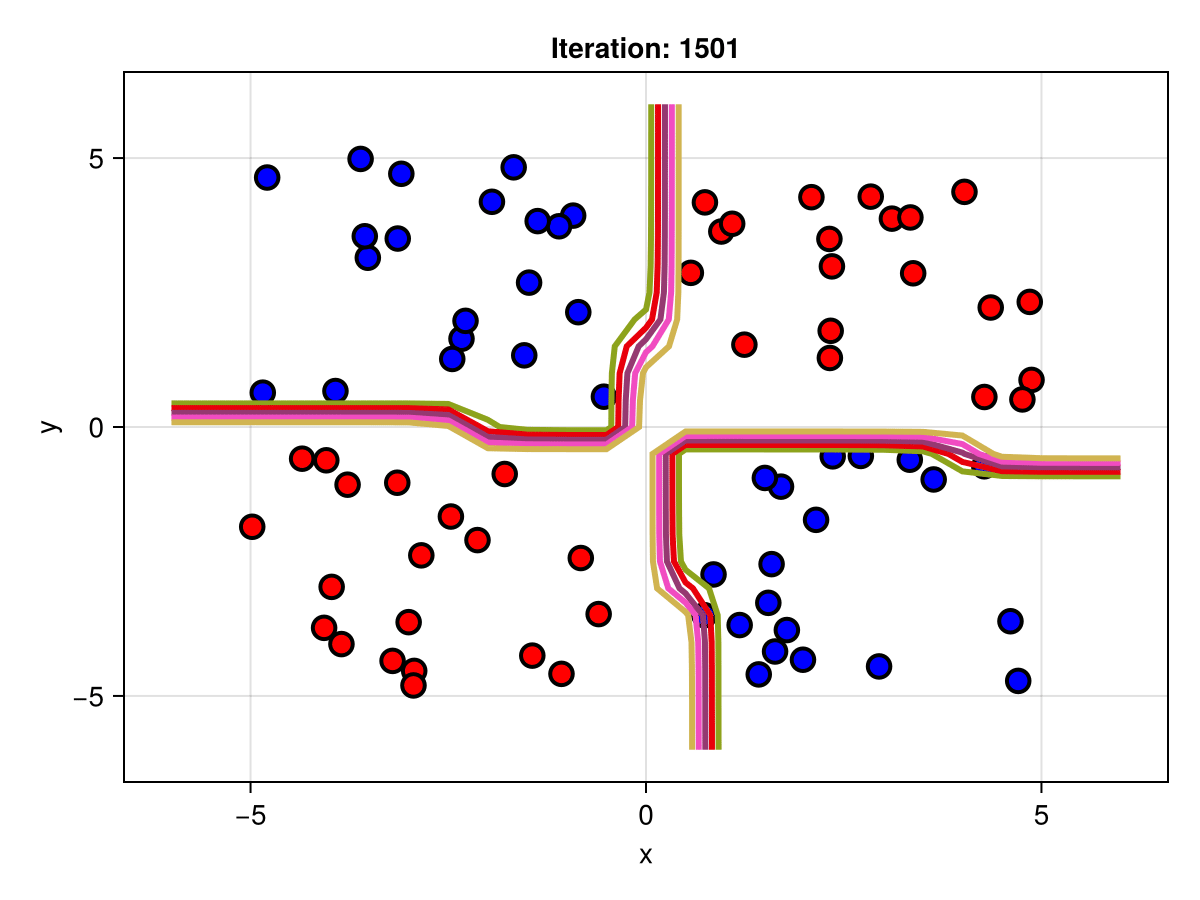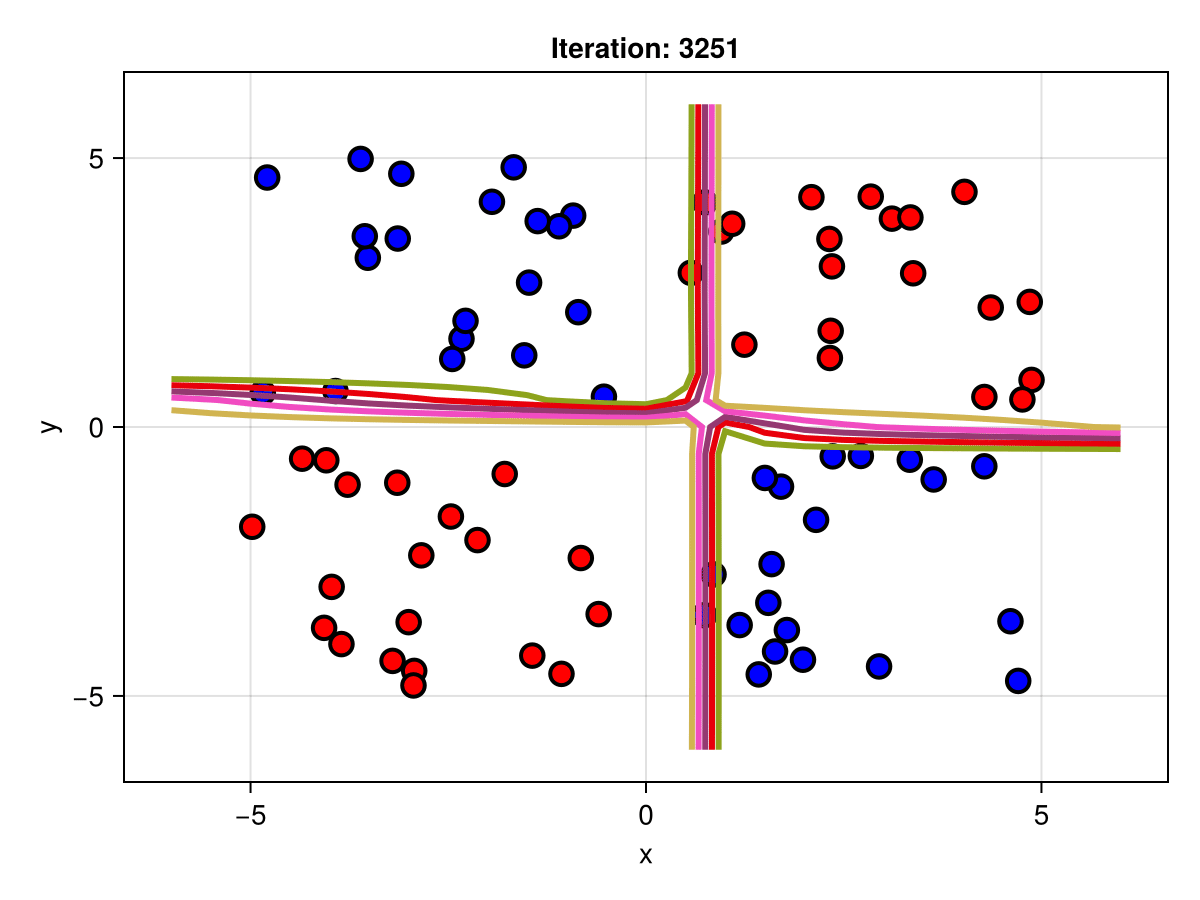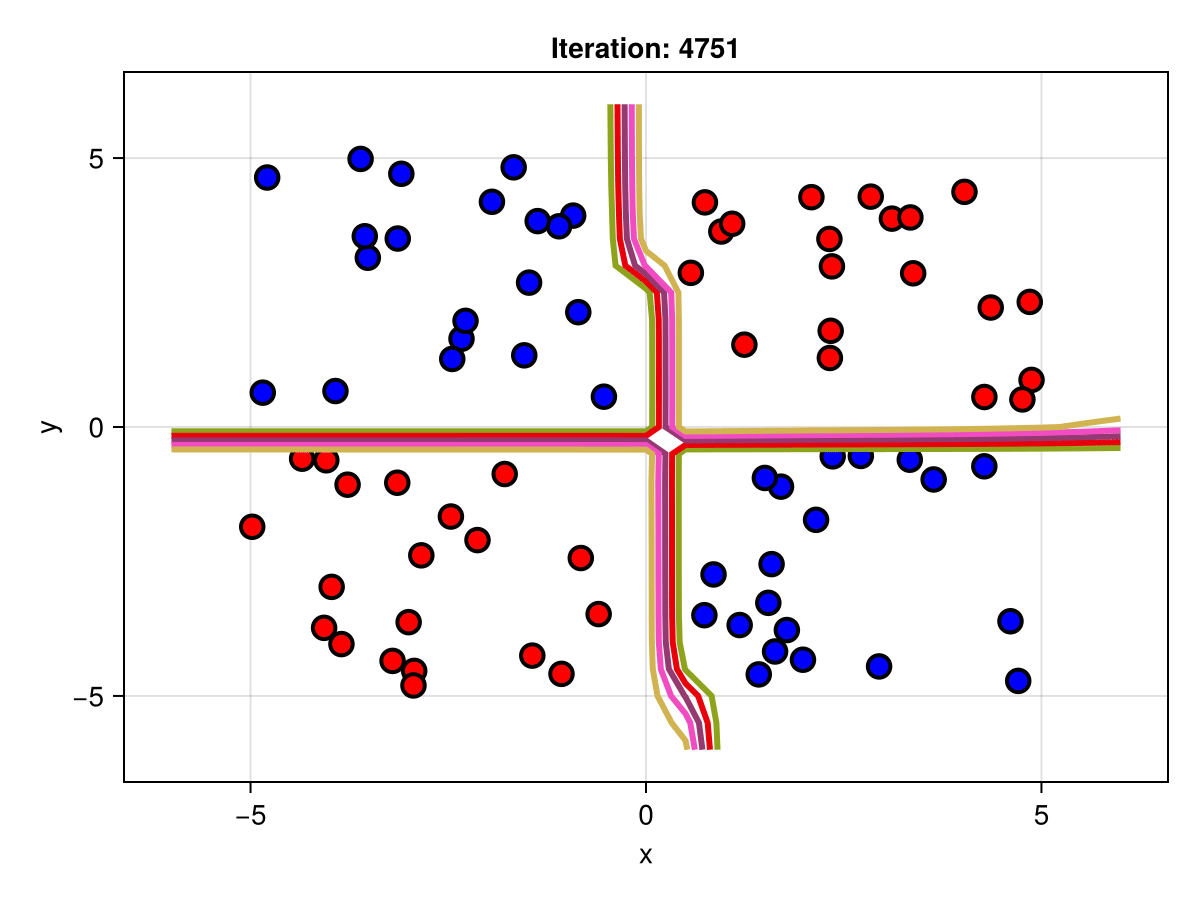
Appendix
using InteractiveUtils
InteractiveUtils.versioninfo()
if @isdefined(MLDataDevices)
if @isdefined(CUDA) && MLDataDevices.functional(CUDADevice)
println()
CUDA.versioninfo()
end
if @isdefined(AMDGPU) && MLDataDevices.functional(AMDGPUDevice)
println()
AMDGPU.versioninfo()
end
endJulia Version 1.11.2
Commit 5e9a32e7af2 (2024-12-01 20:02 UTC)
Build Info:
Official https://julialang.org/ release
Platform Info:
OS: Linux (x86_64-linux-gnu)
CPU: 128 × AMD EPYC 7502 32-Core Processor
WORD_SIZE: 64
LLVM: libLLVM-16.0.6 (ORCJIT, znver2)
Threads: 16 default, 0 interactive, 8 GC (on 16 virtual cores)
Environment:
JULIA_CPU_THREADS = 16
JULIA_DEPOT_PATH = /cache/julia-buildkite-plugin/depots/01872db4-8c79-43af-ab7d-12abac4f24f6
JULIA_PKG_SERVER =
JULIA_NUM_THREADS = 16
JULIA_CUDA_HARD_MEMORY_LIMIT = 100%
JULIA_PKG_PRECOMPILE_AUTO = 0
JULIA_DEBUG = LiterateThis page was generated using Literate.jl.