Visualizing Lux Models using Model Explorer
We can use model explorer to visualize both Lux models and the corresponding gradient expressions. To do this we just need to compile our model using Reactant and save the resulting mlir file.
julia
using Lux, Reactant, Enzyme, Random
dev = reactant_device(; force=true)
model = Chain(
Chain(
Conv((3, 3), 3 => 32, relu; pad=SamePad()),
BatchNorm(32),
),
FlattenLayer(),
Dense(32 * 32 * 32 => 32, tanh),
BatchNorm(32),
Dense(32 => 10)
)
ps, st = Lux.setup(Random.default_rng(), model) |> dev
x = randn(Float32, 32, 32, 3, 4) |> devFollowing instructions from exporting lux models to stablehlo we can save the mlir file.
julia
hlo = @code_hlo model(x, ps, Lux.testmode(st))
write("exported_lux_model.mlir", string(hlo))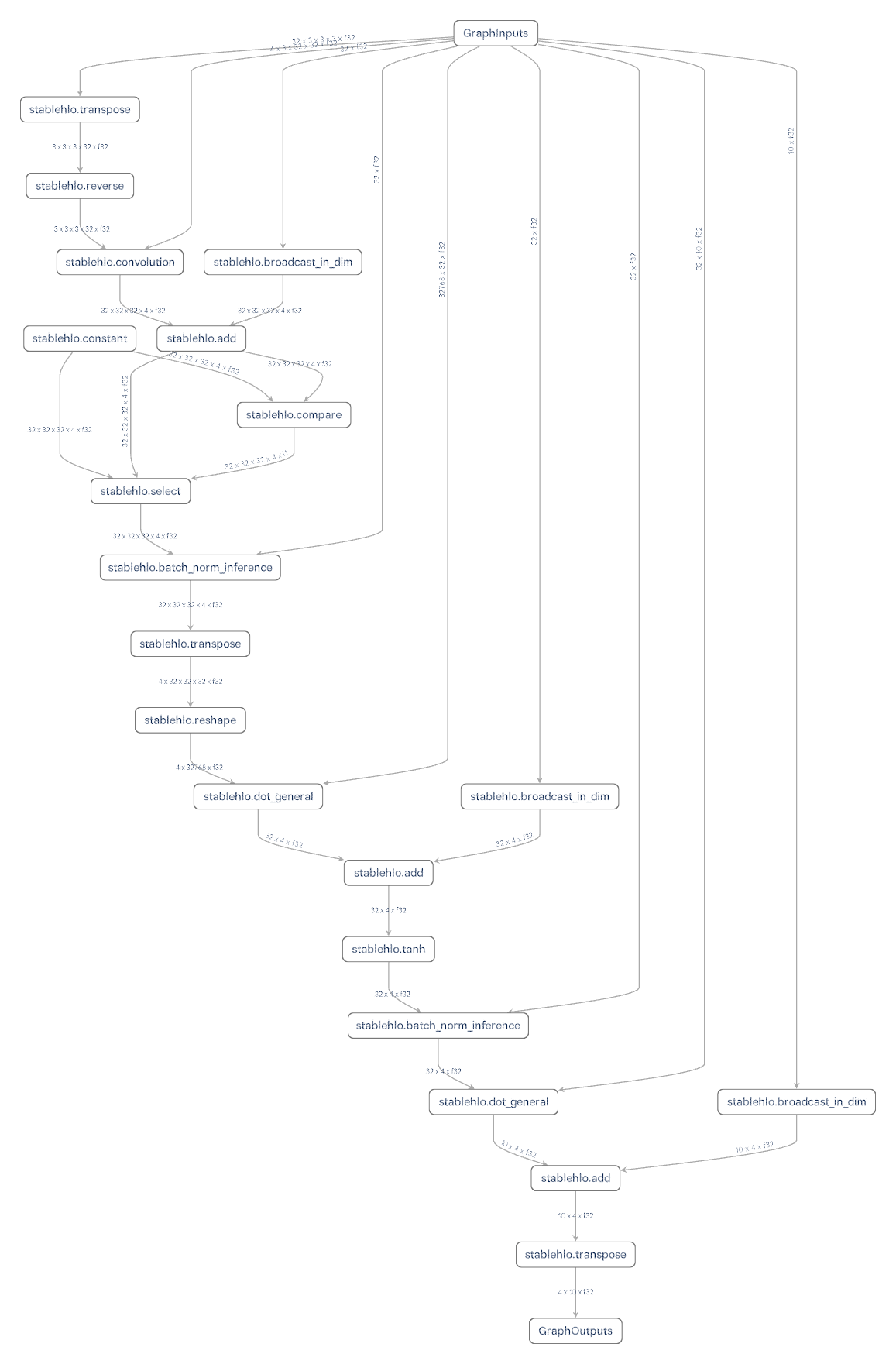
We can also visualize the gradients of the model using the same method.
julia
function ∇sumabs2_enzyme(model, x, ps, st)
return Enzyme.gradient(Enzyme.Reverse, sum ∘ first ∘ Lux.apply, Const(model),
x, ps, Const(st))
end
hlo = @code_hlo ∇sumabs2_enzyme(model, x, ps, st)
write("exported_lux_model_gradients.mlir", string(hlo))This is going to be hard to read, but you get the idea.
