Not Run on CI
This tutorial is not run on CI to reduce the computational burden. If you encounter any issues, please open an issue on the Lux.jl repository.
Normalizing Flows for Density Estimation
This tutorial demonstrates how to use Lux to train a RealNVP. This is based on the RealNVP implementation in MLX.
julia
using Lux,
Reactant,
Random,
Statistics,
Enzyme,
MLUtils,
ConcreteStructs,
Printf,
Optimisers,
CairoMakie
const xdev = reactant_device(; force=true)
const cdev = cpu_device()Define & Load the Moons Dataset
We define a function to generate data from the moons dataset. We use the code here from this tutorial.
julia
function make_moons(
rng::AbstractRNG,
::Type{T},
n_samples::Int=100;
noise::Union{Nothing,AbstractFloat}=nothing,
) where {T}
n_moons = n_samples ÷ 2
t_min, t_max = T(0), T(π)
t_inner = rand(rng, T, n_moons) * (t_max - t_min) .+ t_min
t_outer = rand(rng, T, n_moons) * (t_max - t_min) .+ t_min
outer_circ_x = cos.(t_outer)
outer_circ_y = sin.(t_outer) .+ T(1)
inner_circ_x = 1 .- cos.(t_inner)
inner_circ_y = 1 .- sin.(t_inner) .- T(1)
data = [outer_circ_x outer_circ_y; inner_circ_x inner_circ_y]
z = permutedims(data, (2, 1))
noise !== nothing && (z .+= T(noise) * randn(rng, T, size(z)))
return z
endLet's visualize the dataset
julia
fig = Figure()
ax = CairoMakie.Axis(fig[1, 1]; xlabel="x", ylabel="y")
z = make_moons(Random.default_rng(), Float32, 10_000; noise=0.1)
scatter!(ax, z[1, :], z[2, :]; markersize=2)
figjulia
function load_moons_dataloader(
args...; batchsize::Int, noise::Union{Nothing,AbstractFloat}=nothing, kwargs...
)
return DataLoader(
make_moons(args...; noise); batchsize, shuffle=true, partial=false, kwargs...
)
endBijectors Implementation
julia
abstract type AbstractBijector end
@concrete struct AffineBijector <: AbstractBijector
shift <: AbstractArray
log_scale <: AbstractArray
end
function AffineBijector(shift_and_log_scale::AbstractArray{T,N}) where {T,N}
n = size(shift_and_log_scale, 1) ÷ 2
idxs = ntuple(Returns(Colon()), N - 1)
return AffineBijector(
shift_and_log_scale[1:n, idxs...], shift_and_log_scale[(n + 1):end, idxs...]
)
end
function forward_and_log_det(bj::AffineBijector, x::AbstractArray)
y = x .* exp.(bj.log_scale) .+ bj.shift
return y, bj.log_scale
end
function inverse_and_log_det(bj::AffineBijector, y::AbstractArray)
x = (y .- bj.shift) ./ exp.(bj.log_scale)
return x, -bj.log_scale
end
@concrete struct MaskedCoupling <: AbstractBijector
mask <: AbstractArray
conditioner
bijector
end
function apply_mask(bj::MaskedCoupling, x::AbstractArray, fn::F) where {F}
x_masked = x .* (1 .- bj.mask)
bijector_params = bj.conditioner(x_masked)
y, log_det = fn(bijector_params)
log_det = log_det .* bj.mask
y = ifelse.(bj.mask, y, x)
return y, dsum(log_det; dims=Tuple(collect(1:(ndims(x) - 1))))
end
function forward_and_log_det(bj::MaskedCoupling, x::AbstractArray)
return apply_mask(bj, x, params -> forward_and_log_det(bj.bijector(params), x))
end
function inverse_and_log_det(bj::MaskedCoupling, y::AbstractArray)
return apply_mask(bj, y, params -> inverse_and_log_det(bj.bijector(params), y))
endModel Definition
julia
function MLP(in_dims::Int, hidden_dims::Int, out_dims::Int, n_layers::Int; activation=gelu)
return Chain(
Dense(in_dims => hidden_dims, activation),
[Dense(hidden_dims => hidden_dims, activation) for _ in 1:(n_layers - 1)]...,
Dense(hidden_dims => out_dims),
)
end
@concrete struct RealNVP <: AbstractLuxContainerLayer{(:conditioners,)}
conditioners
dist_dims::Int
n_transforms::Int
end
const StatefulRealNVP{M} = StatefulLuxLayer{M,<:RealNVP}
function Lux.initialstates(rng::AbstractRNG, l::RealNVP)
mask_list = Vector{Bool}[
collect(1:(l.dist_dims)) .% 2 .== i % 2 for i in 1:(l.n_transforms)
]
return (; mask_list, conditioners=Lux.initialstates(rng, l.conditioners))
end
function RealNVP(; n_transforms::Int, dist_dims::Int, hidden_dims::Int, n_layers::Int)
conditioners = [
MLP(dist_dims, hidden_dims, 2 * dist_dims, n_layers; activation=gelu) for
_ in 1:n_transforms
]
conditioners = NamedTuple{ntuple(Base.Fix1(Symbol, :conditioners_), n_transforms)}(
Tuple(conditioners)
)
return RealNVP(conditioners, dist_dims, n_transforms)
end
log_prob(x::AbstractArray{T}) where {T} = -T(0.5 * log(2π)) .- T(0.5) .* abs2.(x)
function log_prob(l::StatefulRealNVP, x::AbstractArray{T}) where {T}
smodels = [
StatefulLuxLayer(conditioner, l.ps.conditioners[i], l.st.conditioners[i]) for
(i, conditioner) in enumerate(l.model.conditioners)
]
lprob = zeros_like(x, size(x, ndims(x)))
for (mask, conditioner) in Iterators.reverse(zip(l.st.mask_list, smodels))
bj = MaskedCoupling(mask, conditioner, AffineBijector)
x, log_det = inverse_and_log_det(bj, x)
lprob += log_det
end
lprob += dsum(log_prob(x); dims=Tuple(collect(1:(ndims(x) - 1))))
conditioners = NamedTuple{
ntuple(Base.Fix1(Symbol, :conditioners_), l.model.n_transforms)
}(
Tuple([smodel.st for smodel in smodels])
)
l.st = merge(l.st, (; conditioners))
return lprob
end
function sample(
rng::AbstractRNG,
::Type{T},
d::StatefulRealNVP,
nsamples::Int,
nsteps::Int=length(d.model.conditioners),
) where {T}
@assert 1 ≤ nsteps ≤ length(d.model.conditioners)
smodels = [
StatefulLuxLayer(conditioner, d.ps.conditioners[i], d.st.conditioners[i]) for
(i, conditioner) in enumerate(d.model.conditioners)
]
x = randn(rng, T, d.model.dist_dims, nsamples)
for (i, (mask, conditioner)) in enumerate(zip(d.st.mask_list, smodels))
x, _ = forward_and_log_det(MaskedCoupling(mask, conditioner, AffineBijector), x)
i ≥ nsteps && break
end
return x
endHelper Functions
julia
dsum(x; dims) = dropdims(sum(x; dims); dims)
function loss_function(model, ps, st, x)
smodel = StatefulLuxLayer(model, ps, st)
lprob = log_prob(smodel, x)
return -mean(lprob), smodel.st, (;)
endTraining the Model
julia
function main(;
maxiters::Int=10_000,
n_train_samples::Int=100_000,
batchsize::Int=128,
n_transforms::Int=6,
hidden_dims::Int=16,
n_layers::Int=4,
lr::Float64=0.0004,
noise::Float64=0.06,
)
rng = Random.default_rng()
Random.seed!(rng, 0)
dataloader =
load_moons_dataloader(rng, Float32, n_train_samples; batchsize, noise) |>
xdev |>
Iterators.cycle
model = RealNVP(; n_transforms, dist_dims=2, hidden_dims, n_layers)
ps, st = Lux.setup(rng, model) |> xdev
opt = Adam(lr)
train_state = Training.TrainState(model, ps, st, opt)
@printf "Total Trainable Parameters: %d\n" Lux.parameterlength(ps)
total_samples = 0
start_time = time()
for (iter, x) in enumerate(dataloader)
total_samples += size(x, ndims(x))
(_, loss, _, train_state) = Training.single_train_step!(
AutoEnzyme(), loss_function, x, train_state; return_gradients=Val(false)
)
isnan(loss) && error("NaN loss encountered in iter $(iter)!")
if iter == 1 || iter == maxiters || iter % 1000 == 0
throughput = total_samples / (time() - start_time)
@printf "Iter: [%6d/%6d]\tTraining Loss: %.6f\t\
Throughput: %.6f samples/s\n" iter maxiters loss throughput
end
iter ≥ maxiters && break
end
return StatefulLuxLayer(model, train_state.parameters, Lux.testmode(train_state.states))
end
trained_model = main()Visualizing the Results
julia
z_stages = Matrix{Float32}[]
for i in 1:(trained_model.model.n_transforms)
z = @jit sample(Random.default_rng(), Float32, trained_model, 10_000, i)
push!(z_stages, Array(z))
end
begin
fig = Figure(; size=(1200, 800))
for (idx, z) in enumerate(z_stages)
i, j = (idx - 1) ÷ 3, (idx - 1) % 3
ax = Axis(fig[i, j]; title="$(idx) transforms")
scatter!(ax, z[1, :], z[2, :]; markersize=2)
end
fig
end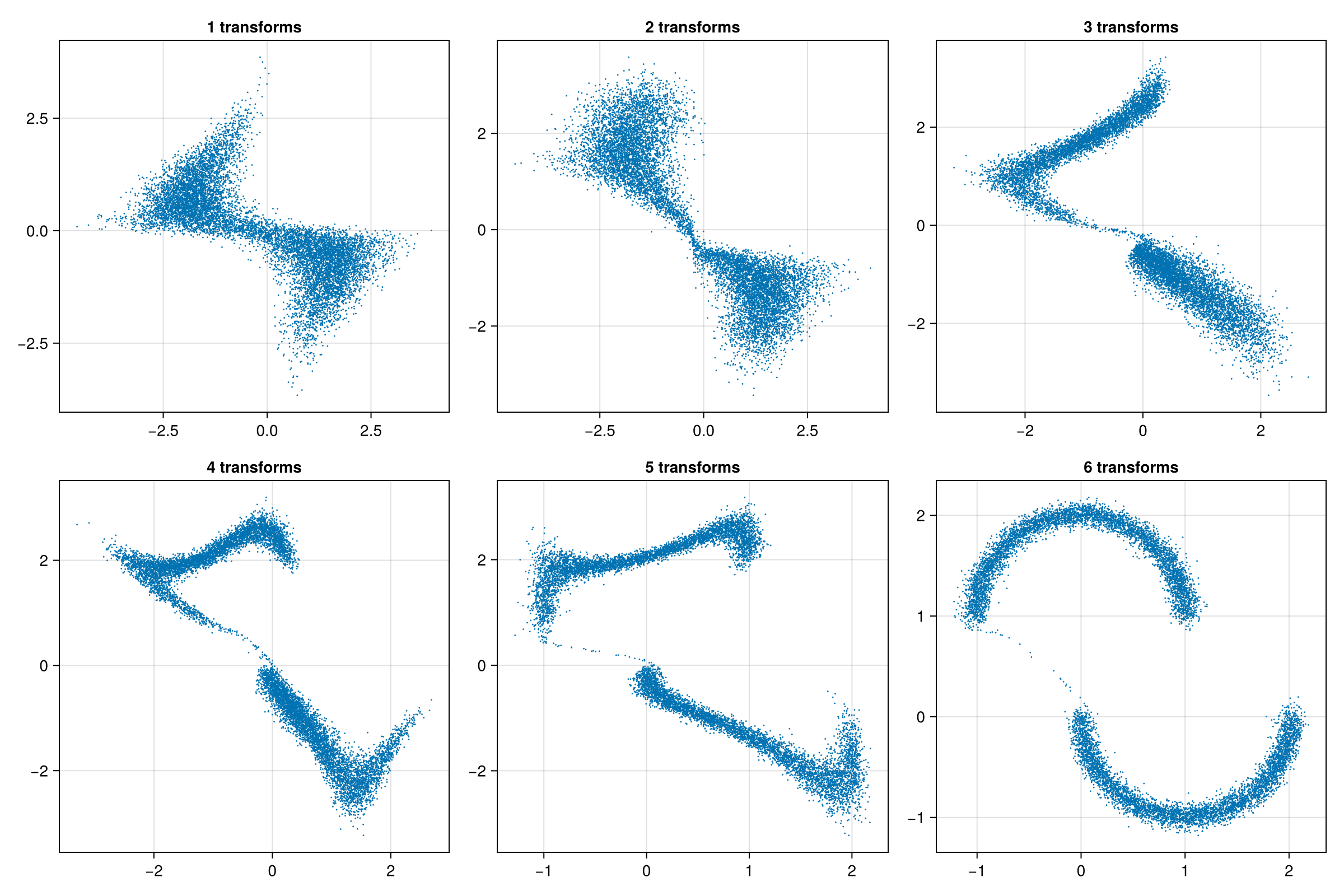
This page was generated using Literate.jl.