Bayesian Neural Network
We borrow this tutorial from the official Turing Docs. We will show how the explicit parameterization of Lux enables first-class composability with packages which expect flattened out parameter vectors.
We will use Turing.jl with Lux.jl to implement implementing a classification algorithm. Lets start by importing the relevant libraries.
# Import libraries
using Lux, Turing, CairoMakie, Random, Tracker, Functors, LinearAlgebra
# Sampling progress
Turing.setprogress!(true);[ Info: [Turing]: progress logging is enabled globally
[ Info: [AdvancedVI]: global PROGRESS is set as trueGenerating data
Our goal here is to use a Bayesian neural network to classify points in an artificial dataset. The code below generates data points arranged in a box-like pattern and displays a graph of the dataset we'll be working with.
# Number of points to generate
N = 80
M = round(Int, N / 4)
rng = Random.default_rng()
Random.seed!(rng, 1234)
# Generate artificial data
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
xt1s = Array([[x1s[i] + 0.5f0; x2s[i] + 0.5f0] for i in 1:M])
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
append!(xt1s, Array([[x1s[i] - 5.0f0; x2s[i] - 5.0f0] for i in 1:M]))
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
xt0s = Array([[x1s[i] + 0.5f0; x2s[i] - 5.0f0] for i in 1:M])
x1s = rand(rng, Float32, M) * 4.5f0;
x2s = rand(rng, Float32, M) * 4.5f0;
append!(xt0s, Array([[x1s[i] - 5.0f0; x2s[i] + 0.5f0] for i in 1:M]))
# Store all the data for later
xs = [xt1s; xt0s]
ts = [ones(2 * M); zeros(2 * M)]
# Plot data points
function plot_data()
x1 = first.(xt1s)
y1 = last.(xt1s)
x2 = first.(xt0s)
y2 = last.(xt0s)
fig = Figure()
ax = CairoMakie.Axis(fig[1, 1]; xlabel="x", ylabel="y")
scatter!(ax, x1, y1; markersize=16, color=:red, strokecolor=:black, strokewidth=2)
scatter!(ax, x2, y2; markersize=16, color=:blue, strokecolor=:black, strokewidth=2)
return fig
end
plot_data()Building the Neural Network
The next step is to define a feedforward neural network where we express our parameters as distributions, and not single points as with traditional neural networks. For this we will use Dense to define liner layers and compose them via Chain, both are neural network primitives from Lux. The network nn we will create will have two hidden layers with tanh activations and one output layer with sigmoid activation, as shown below.
The nn is an instance that acts as a function and can take data, parameters and current state as inputs and output predictions. We will define distributions on the neural network parameters.
# Construct a neural network using Lux
nn = Chain(Dense(2 => 3, tanh), Dense(3 => 2, tanh), Dense(2 => 1, sigmoid))
# Initialize the model weights and state
ps, st = Lux.setup(rng, nn)
Lux.parameterlength(nn) # number of paraemters in NN20The probabilistic model specification below creates a parameters variable, which has IID normal variables. The parameters represents all parameters of our neural net (weights and biases).
# Create a regularization term and a Gaussian prior variance term.
alpha = 0.09
sig = sqrt(1.0 / alpha)3.3333333333333335Construct named tuple from a sampled parameter vector. We could also use ComponentArrays here and simply broadcast to avoid doing this. But let's do it this way to avoid dependencies.
function vector_to_parameters(ps_new::AbstractVector, ps::NamedTuple)
@assert length(ps_new) == Lux.parameterlength(ps)
i = 1
function get_ps(x)
z = reshape(view(ps_new, i:(i + length(x) - 1)), size(x))
i += length(x)
return z
end
return fmap(get_ps, ps)
endvector_to_parameters (generic function with 1 method)To interface with external libraries it is often desirable to use the StatefulLuxLayer to automatically handle the neural network states.
const model = StatefulLuxLayer(nn, st)
# Specify the probabilistic model.
@model function bayes_nn(xs, ts)
# Sample the parameters
nparameters = Lux.parameterlength(nn)
parameters ~ MvNormal(zeros(nparameters), Diagonal(abs2.(sig .* ones(nparameters))))
# Forward NN to make predictions
preds = Lux.apply(model, xs, vector_to_parameters(parameters, ps))
# Observe each prediction.
for i in eachindex(ts)
ts[i] ~ Bernoulli(preds[i])
end
endbayes_nn (generic function with 2 methods)Inference can now be performed by calling sample. We use the HMC sampler here.
# Perform inference.
N = 5000
ch = sample(bayes_nn(reduce(hcat, xs), ts), HMC(0.05, 4; adtype=AutoTracker()), N)Chains MCMC chain (5000×30×1 Array{Float64, 3}):
Iterations = 1:1:5000
Number of chains = 1
Samples per chain = 5000
Wall duration = 24.99 seconds
Compute duration = 24.99 seconds
parameters = parameters[1], parameters[2], parameters[3], parameters[4], parameters[5], parameters[6], parameters[7], parameters[8], parameters[9], parameters[10], parameters[11], parameters[12], parameters[13], parameters[14], parameters[15], parameters[16], parameters[17], parameters[18], parameters[19], parameters[20]
internals = lp, n_steps, is_accept, acceptance_rate, log_density, hamiltonian_energy, hamiltonian_energy_error, numerical_error, step_size, nom_step_size
Summary Statistics
parameters mean std mcse ess_bulk ess_tail rhat ess_per_sec
Symbol Float64 Float64 Float64 Float64 Float64 Float64 Float64
parameters[1] -0.5133 1.8835 0.5330 13.1888 25.0326 1.4853 0.5277
parameters[2] -5.3361 2.4104 0.6377 15.5139 36.9602 1.0376 0.6208
parameters[3] 0.3151 0.6835 0.1441 27.6358 50.3442 1.1216 1.1058
parameters[4] 1.9624 3.7648 1.1478 11.5629 25.5317 2.0118 0.4627
parameters[5] -0.0914 0.6013 0.0858 52.3129 54.6520 1.1223 2.0932
parameters[6] 4.9348 2.3882 0.6909 12.4651 22.5599 1.8704 0.4988
parameters[7] -1.6494 2.7707 0.8306 11.9136 35.7353 1.9059 0.4767
parameters[8] -0.3367 1.5561 0.3957 15.3027 35.5527 1.2732 0.6123
parameters[9] -0.6247 1.7715 0.4439 16.1582 41.0067 1.0886 0.6465
parameters[10] -0.0485 2.6496 0.7711 12.0002 18.2390 1.6173 0.4802
parameters[11] -2.7777 2.4100 0.6937 13.2493 54.0054 1.2589 0.5301
parameters[12] 1.7397 2.7286 0.8144 12.5350 30.6674 1.3093 0.5016
parameters[13] -1.3065 3.0122 0.9212 11.3718 29.5307 1.8415 0.4550
parameters[14] 2.1359 1.7145 0.4376 16.0168 29.9135 1.1185 0.6409
parameters[15] -2.2917 1.1075 0.2143 27.1032 53.9717 1.0339 1.0845
parameters[16] -1.8241 1.7783 0.4754 14.8255 44.7465 1.2380 0.5932
parameters[17] -2.9541 1.0941 0.1964 32.4857 49.3634 1.0550 1.2998
parameters[18] -2.9283 3.1083 0.9424 13.4337 49.1159 1.4880 0.5375
parameters[19] -5.8506 1.2702 0.1981 41.3283 64.0949 1.0032 1.6537
parameters[20] -3.6909 1.8615 0.5266 13.1481 52.3750 1.6064 0.5261
Quantiles
parameters 2.5% 25.0% 50.0% 75.0% 97.5%
Symbol Float64 Float64 Float64 Float64 Float64
parameters[1] -2.9584 -2.1586 -0.3884 0.5613 4.2684
parameters[2] -9.8741 -7.2592 -4.9623 -3.2715 -1.7035
parameters[3] -0.6888 -0.1073 0.1721 0.5713 2.2193
parameters[4] -5.7669 -1.4891 3.0249 5.1736 7.8110
parameters[5] -1.2037 -0.4726 -0.1166 0.2270 1.2686
parameters[6] 1.5300 2.9517 4.4678 6.7396 10.5049
parameters[7] -5.8618 -4.2435 -1.3364 0.6954 3.0399
parameters[8] -3.2162 -1.3171 -0.3432 0.4455 3.4264
parameters[9] -4.2319 -1.6828 -0.5051 0.6070 2.9411
parameters[10] -6.1276 -1.8893 0.2035 1.6808 4.6669
parameters[11] -6.8391 -4.9499 -2.5672 -0.5826 1.1874
parameters[12] -3.4081 -0.4600 2.2126 3.9338 5.8529
parameters[13] -6.1325 -3.7644 -2.1160 1.6571 3.6084
parameters[14] -1.1205 1.0660 1.9677 3.1250 5.5755
parameters[15] -4.6287 -3.0619 -2.1911 -1.5340 -0.1136
parameters[16] -5.3259 -3.0340 -1.9008 -0.9440 1.7702
parameters[17] -5.6472 -3.5470 -2.8160 -2.2108 -1.0283
parameters[18] -6.7277 -5.2922 -4.2915 0.7602 2.7335
parameters[19] -8.5012 -6.5957 -5.8670 -5.0328 -3.3450
parameters[20] -6.7333 -5.1577 -3.8273 -2.2208 -0.3195Now we extract the parameter samples from the sampled chain as θ (this is of size 5000 x 20 where 5000 is the number of iterations and 20 is the number of parameters). We'll use these primarily to determine how good our model's classifier is.
# Extract all weight and bias parameters.
θ = MCMCChains.group(ch, :parameters).value;Prediction Visualization
# A helper to run the nn through data `x` using parameters `θ`
nn_forward(x, θ) = model(x, vector_to_parameters(θ, ps))
# Plot the data we have.
fig = plot_data()
# Find the index that provided the highest log posterior in the chain.
_, i = findmax(ch[:lp])
# Extract the max row value from i.
i = i.I[1]
# Plot the posterior distribution with a contour plot
x1_range = collect(range(-6; stop=6, length=25))
x2_range = collect(range(-6; stop=6, length=25))
Z = [nn_forward([x1, x2], θ[i, :])[1] for x1 in x1_range, x2 in x2_range]
contour!(x1_range, x2_range, Z; linewidth=3, colormap=:seaborn_bright)
figThe contour plot above shows that the MAP method is not too bad at classifying our data. Now we can visualize our predictions.
The nn_predict function takes the average predicted value from a network parameterized by weights drawn from the MCMC chain.
# Return the average predicted value across multiple weights.
nn_predict(x, θ, num) = mean([first(nn_forward(x, view(θ, i, :))) for i in 1:10:num])nn_predict (generic function with 1 method)Next, we use the nn_predict function to predict the value at a sample of points where the x1 and x2 coordinates range between -6 and 6. As we can see below, we still have a satisfactory fit to our data, and more importantly, we can also see where the neural network is uncertain about its predictions much easier–-those regions between cluster boundaries.
Plot the average prediction.
fig = plot_data()
n_end = 1500
x1_range = collect(range(-6; stop=6, length=25))
x2_range = collect(range(-6; stop=6, length=25))
Z = [nn_predict([x1, x2], θ, n_end)[1] for x1 in x1_range, x2 in x2_range]
contour!(x1_range, x2_range, Z; linewidth=3, colormap=:seaborn_bright)
figSuppose we are interested in how the predictive power of our Bayesian neural network evolved between samples. In that case, the following graph displays an animation of the contour plot generated from the network weights in samples 1 to 5,000.
fig = plot_data()
Z = [first(nn_forward([x1, x2], θ[1, :])) for x1 in x1_range, x2 in x2_range]
c = contour!(x1_range, x2_range, Z; linewidth=3, colormap=:seaborn_bright)
record(fig, "results.gif", 1:250:size(θ, 1)) do i
fig.current_axis[].title = "Iteration: $i"
Z = [first(nn_forward([x1, x2], θ[i, :])) for x1 in x1_range, x2 in x2_range]
c[3] = Z
return fig
end"results.gif"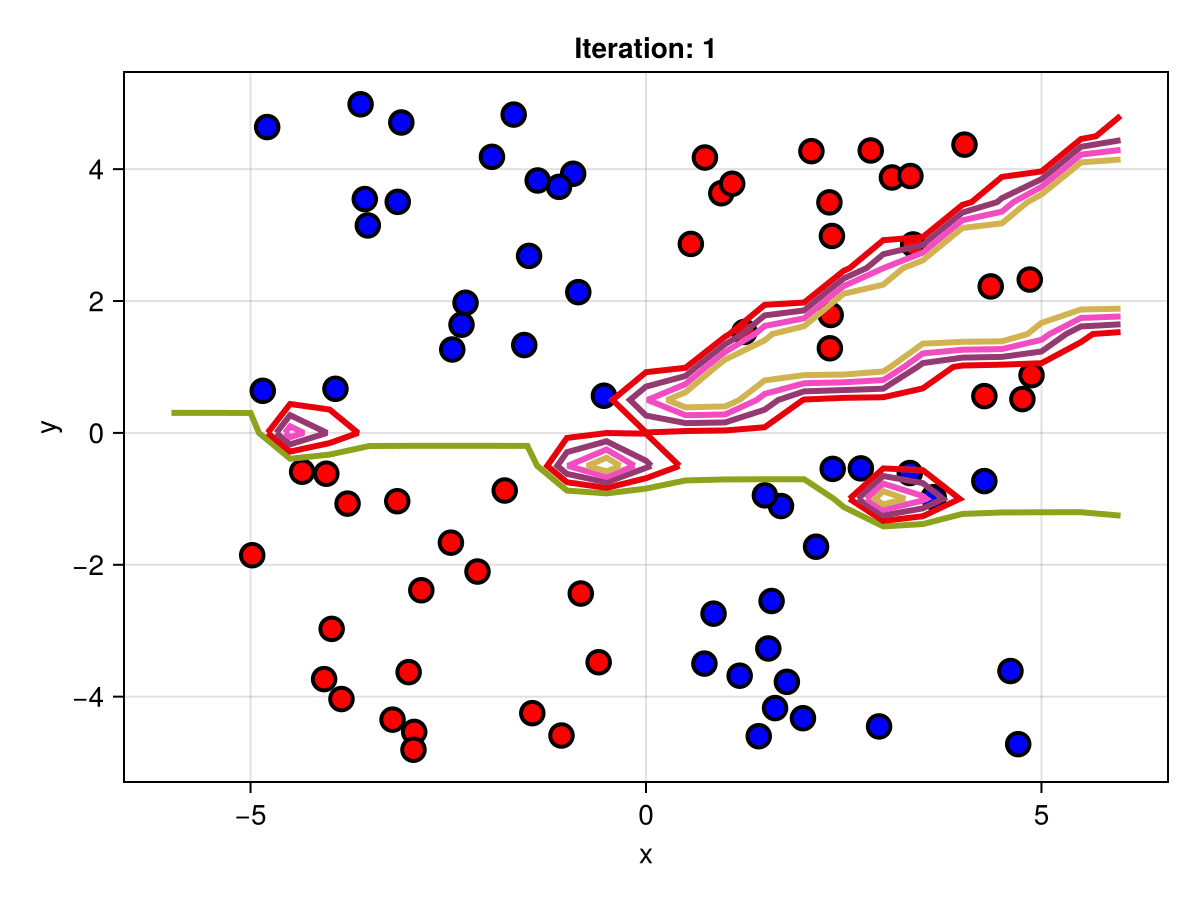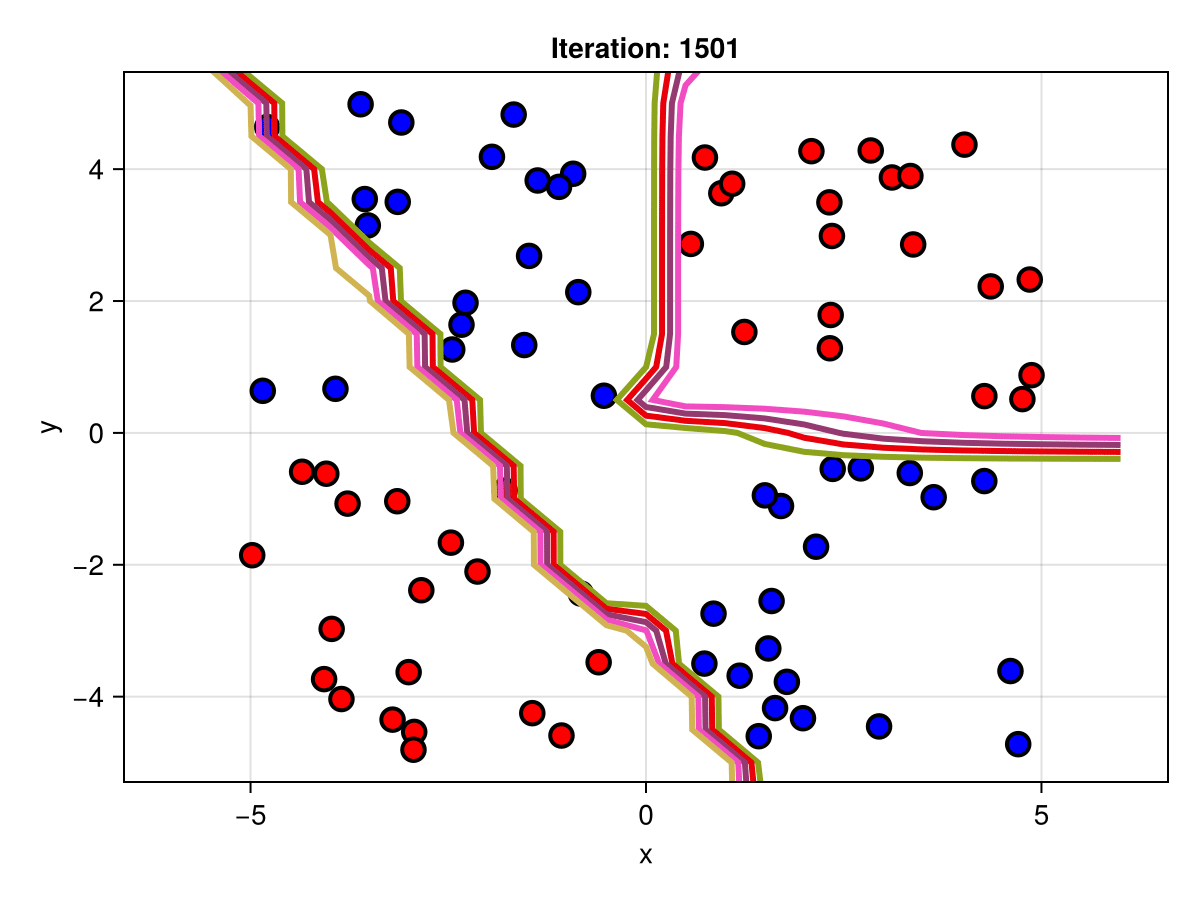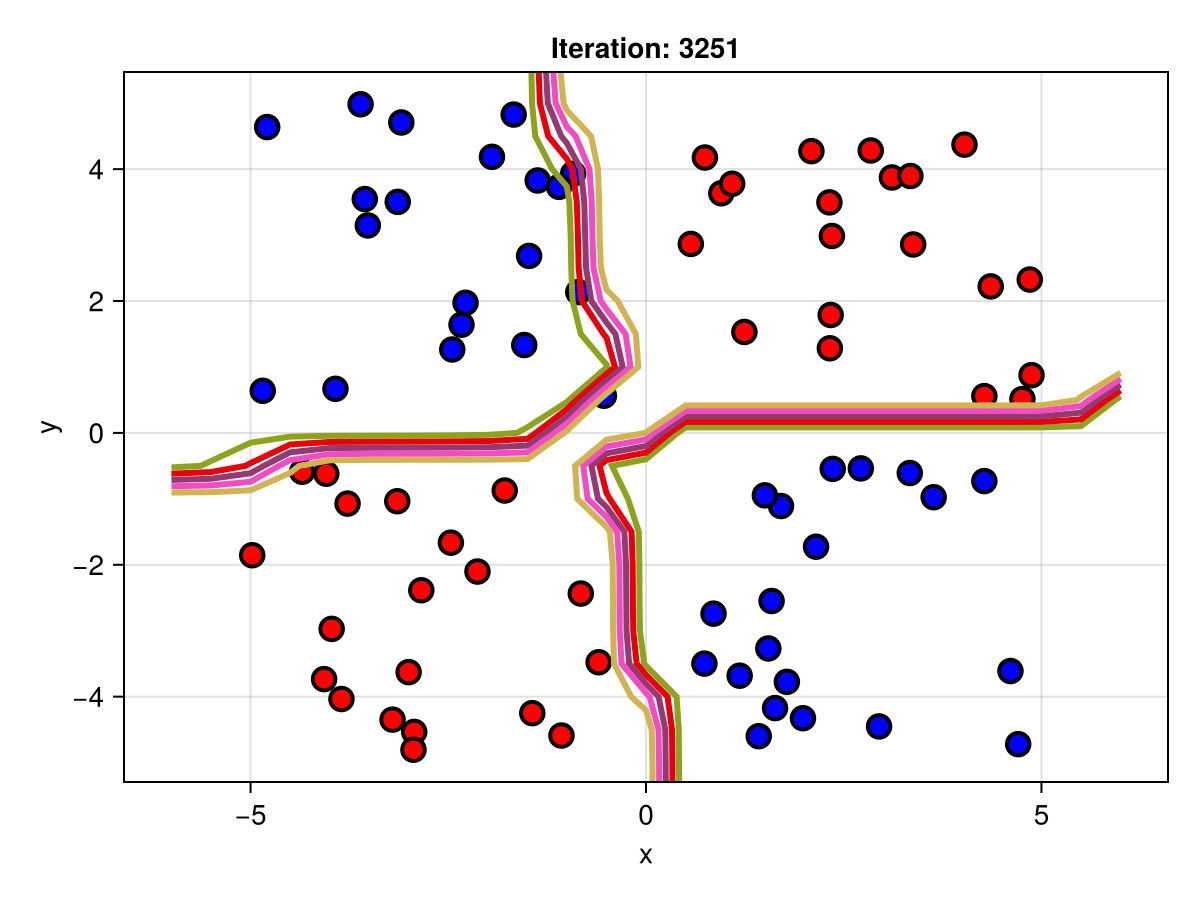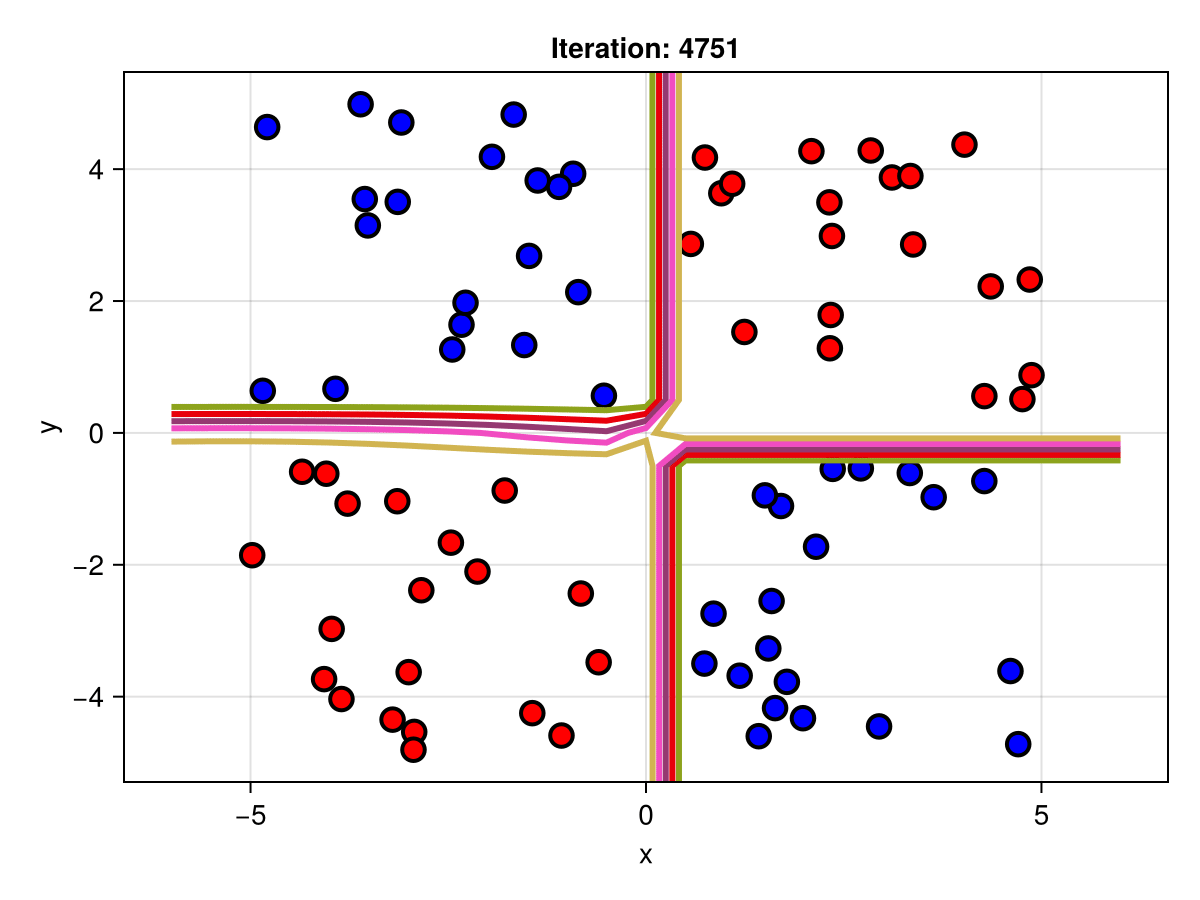
Appendix
using InteractiveUtils
InteractiveUtils.versioninfo()
if @isdefined(LuxCUDA) && CUDA.functional(); println(); CUDA.versioninfo(); end
if @isdefined(LuxAMDGPU) && LuxAMDGPU.functional(); println(); AMDGPU.versioninfo(); endJulia Version 1.10.2
Commit bd47eca2c8a (2024-03-01 10:14 UTC)
Build Info:
Official https://julialang.org/ release
Platform Info:
OS: Linux (x86_64-linux-gnu)
CPU: 48 × AMD EPYC 7402 24-Core Processor
WORD_SIZE: 64
LIBM: libopenlibm
LLVM: libLLVM-15.0.7 (ORCJIT, znver2)
Threads: 48 default, 0 interactive, 24 GC (on 2 virtual cores)
Environment:
LD_LIBRARY_PATH = /usr/local/nvidia/lib:/usr/local/nvidia/lib64
JULIA_DEPOT_PATH = /root/.cache/julia-buildkite-plugin/depots/01872db4-8c79-43af-ab7d-12abac4f24f6
JULIA_PROJECT = /var/lib/buildkite-agent/builds/gpuci-1/julialang/lux-dot-jl/docs
JULIA_AMDGPU_LOGGING_ENABLED = true
JULIA_DEBUG = Literate
JULIA_CPU_THREADS = 2
JULIA_NUM_THREADS = 48
JULIA_LOAD_PATH = @:@v#.#:@stdlib
JULIA_CUDA_HARD_MEMORY_LIMIT = 25%This page was generated using Literate.jl.